CBFBIRN Processing
The Cerebral Blood Flow Biomedical Informatics Research Network (CBFBIRN) was a project to collect and share CBF data acquired using a magnetic resonance imaging (MRI)-based perfusion technique called arterial spin labeling (ASL).
The project’s current iteration consists of several processing scripts that intake p-files and DICOMs to produce BRIKs for further analysis.
Code Flow
To process your data, follows this sequence of execution in MATLAB:
SubjectSetupScript
dofmapGEN
processASLGEN2
Understanding Your Files
We recommend you take a look at your cfmricp_log.txt file to see what each of your files represent. An example is shown below, indicating your scans for the s-folders (DICOMs) and p-files (raw GE scanner data).
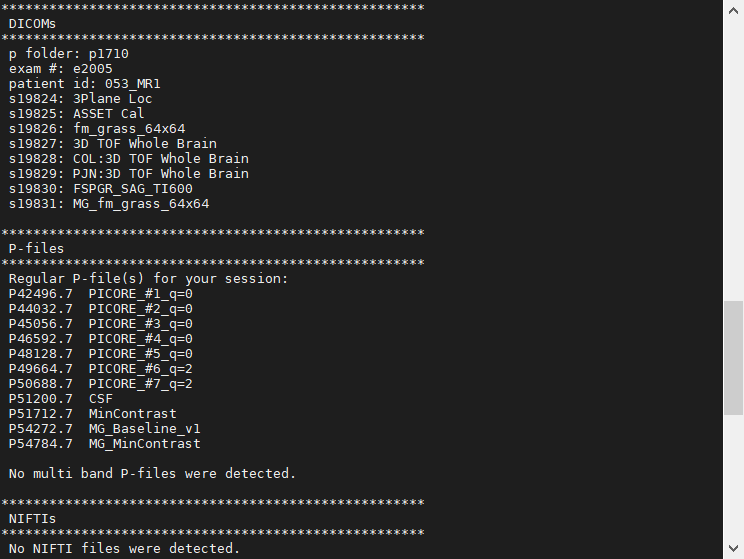
All MRI scans record the raw k-space data acquired by each of the receive coil (at CMFRI, either the 8ch or 32ch) channels during the scan. For many scans, the scanner itself converts this raw k-space data to DICOMs as output. Using certain tools, these DICOMs can be converted to formats like BRIK or NIFTI for research needs. For remaining scans, the raw k-space data is output as a p-file. These are typically “research-grade” scans that need external tools to convert into BRIK or NIFTI formats for further research use.
Subject Setup Script
The subject setup script is a MATLAB file where you specify:
Data directory.
Anatomical and Fieldmap DICOM folders.
ASL, CSF, and MinCon p-files.
Processed directory name.
Name stems for output BRIKs.
Constructing Initial BRIKs and Fieldmap Correction - dofmapGEN
The dofmapGEN script first converts p-files to BRIKs, then applies fieldmap correction to those BRIKs. For example, the ASL data p-file is converted into aslbrik+orig on the first step (the exact name would depend on the previous step’s setup script). After fieldmap correction, the output is called aslbrik+dbl+orig (where +dbl stands for deblurring to indicate fieldmap corrected).
What is the purpose of fieldmap correction? This is an important step that can correct some artifacts in the data. Areas of the head like the sinuses tend to distort the local magnetic field. To the scanner, this causes signals to “look like” they’re coming from one area of the head while in reality they come from an area nearby. With spiral k-space acquisitions, this misattribution tends to cause bunching artifacts in the reconstructed image.
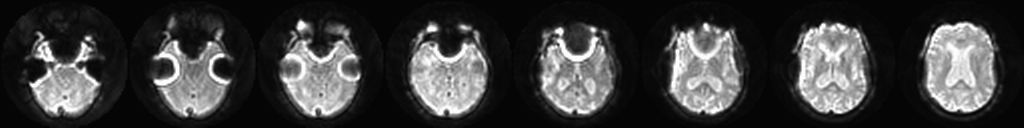
The fieldmap scan measures the magnetic field around the head. Fieldmap correction uses this data to estimate what the image should really look like - to “undo” the artifact as best it can.
Quantifying CBF - processASLGEN2
The processASLGEN2 script completes the quantification of CBF data. Overall, it writes out a number of additional outputs:
ASL data after volume registration
CSF data after alignment to ASL
Anatomical data after alignment to ASL, skull-stripping, partial volume estimation, tissue segmentation
CBF data
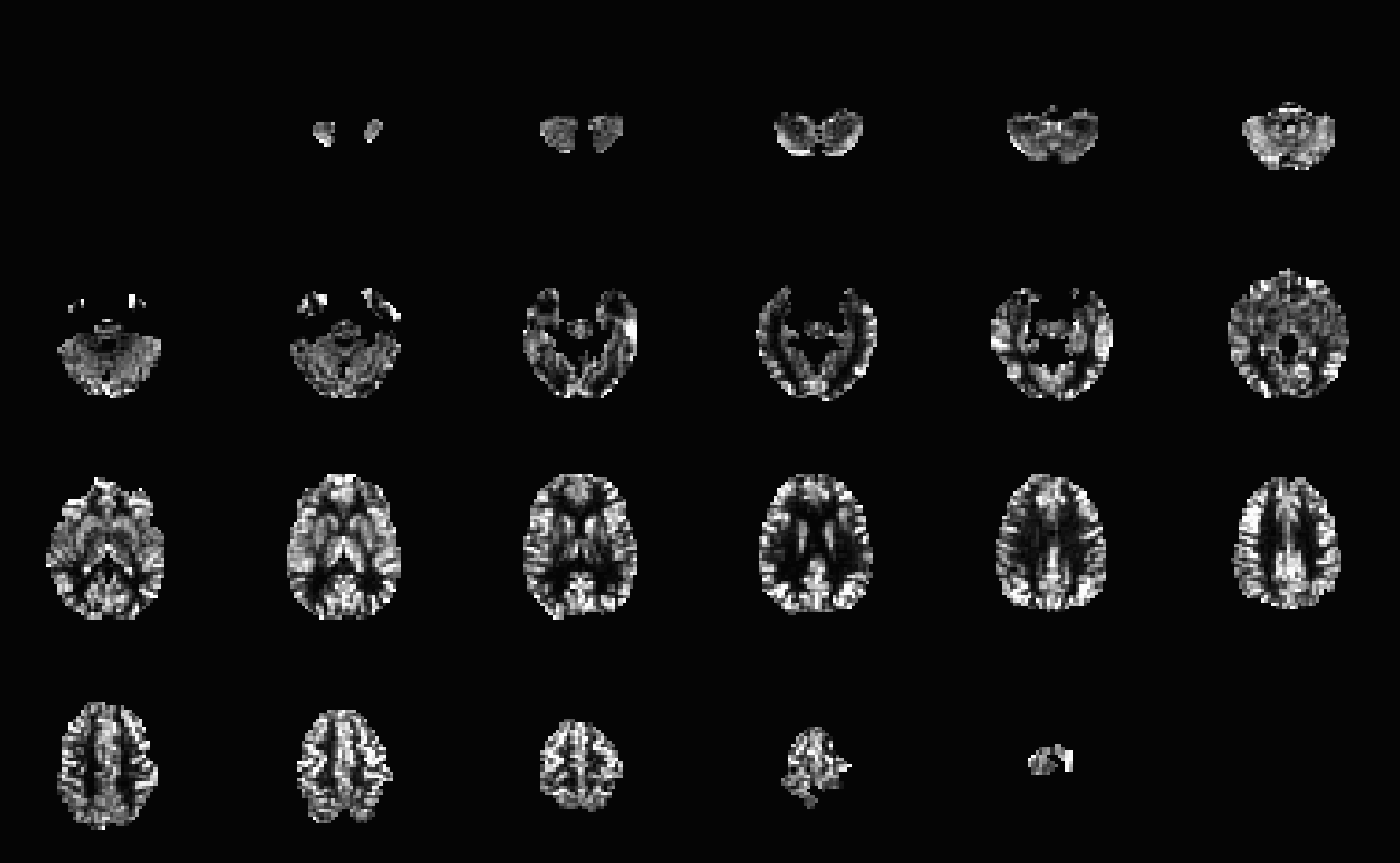
The exact CBF quantification processing depends on many factors, such as whether PASL (pulsed ASL) or PCASL (pseudocontinuous ASL) was used, background suppression was applied, voxel-wise vs reference tissue calibration, etc. Ultimately, the pipeline should CBF images like below.
Reference - Table of Outputs
Fieldmap Correction Outputs
(Scroll sideways to see complete table)
Output |
Description |
|---|---|
csfbrik+orig |
Proton-density image used for calibration of CBF data. This output only exists if the CSF data is acquired separately from the ASL data. |
csfbrik+dbl+orig |
Fieldmap correction applied to csfbrik+orig. This output only exists if the CSF data is acquired separately from the ASL data. |
aslbrik+orig |
BRIK constructed from the ASL scan p-file |
aslbrik+dbl+orig |
Fieldmap correction applied to aslbrik+orig |
aslbrik+dbltcat+orig |
The ASL data from aslbrik+dbl+orig. This ‘tcat’ output only exists if the CSF and ASL data are acquired in the same scan. |
minconbrik+orig |
Minimum contrast data used for coil sensitivity correction. |
minconbrik+dbl+orig |
Minimum contrast data after fieldmap correction. |
ASL Processing Outputs
(Scroll sideways to see complete table)
Output |
Description |
Command |
|---|---|---|
anat+offset+nonRAI+orig |
The anatomical data. |
Custom MATLAB code. |
anat+offset+orig |
The same anatomical data but reoriented to RAI - 1st axis is R(-) to L(+), 2nd axis is A(-) to P(+), 3rd axis is I(-) to S(+). |
Custom MATLAB code. |
anat+orig |
The anatomical data anat+orig refitted to the ASL data by centering their prescriptions. |
Custom MATLAB code. |
aslbrik+dbltcat+reg+orig |
Volume registration applied to the ASL data using AFNI’s 3dvolreg. If CSF and ASL are acquired in the same scan, this will be aslbrik+dbl+reg+orig instead. |
3dvolreg -Fourier -dfile aslbrik+dbltcat+reg+motion -base aslbrik+dbltcat+orig[middlerep#] -prefix aslbrik+dbltcat+reg pcaslGMbrik+dbltcat+orig
|
aslbrik+dbltcat+reg+motion |
Motion parameters from applying 3dvolreg to the ASL data. |
3dvolreg -Fourier -dfile aslbrik+dbltcat+reg+motion -base aslbrik+dbltcat+orig[middlerep#] -prefix aslbrik+dbltcat+reg pcaslGMbrik+dbltcat+orig
|
aslbrik+dbltcat+reg+d3+orig |
Applying d3 filtering (-1/4 +1/2 -1/4) to aslbrik+dbltcat+reg+orig |
Custom MATLAB code. |
aslbrik+dbltcat+reg+d3+mean+orig |
Averaging aslbrik+dbltcat+reg+d3+orig across time. |
Custom MATLAB code. |
csfbriktcat1+orig |
Proton-density image used for calibration of CBF data. This ‘tcat’ output only exists if the CSF and ASL data are acquired in the same scan. |
3dTcat -prefix csfbriktcat 'aslbrik+dbl+orig[endrep#]'
|
csfbriktcat1+reg+orig |
Proton-density image registered to the ASL data. |
align_epi_anat.py -dset1 csfbriktcat1+orig -dset2 aslbrik+dbltcat+reg+orig -dset1to2 -dset2_base middlerep# -master_dset1 aslbrik+dbltcat+reg+orig -dset1_strip None -dset2_strip None -ginormous_move -suffix +reg
|
anat_ns+orig |
The skullstripped anatomical |
3dSkullStrip -input anat+orig -prefix anat_ns
|
anat_ns_al+orig |
The skullstripped anatomical aligned to the CSF data (which was aligned to the ASL data). |
align_epi_anat.py -anat anat_ns+orig -epi csfbriktcat1+reg+orig -epi_base 0 -anat2epi -anat_has_skull no -epi_strip 3dAutomask
|
anat_ns_al_mat.aff12.1D |
The transformation matrix for aligning the skullstripped anatomical to the CSF data. |
align_epi_anat.py -anat anat_ns+orig -epi csfbriktcat1+reg+orig -epi_base 0 -anat2epi -anat_has_skull no -epi_strip 3dAutomask
|
(deprecated) anat+reg_+orig |
The anatomical data anat+orig refitted to the ASL data by centering their prescriptions. |
Custom MATLAB |
(deprecated) anat+reg__ns+orig |
Skull-stripped output of anat+reg_+orig using AFNI’s 3dSkullStrip. |
3dSkullStrip -input anat+reg_+orig -prefix anat+reg__ns
|
(deprecated) anat+reg__ns_al+orig |
Skull-stripped and aligned/registered anatomical data (aligned to the ASL data) using AFNI’s align_epi_anat.py. |
align_epi_anat.py -anat anat+reg__ns+orig -epi csfbriktcat1+reg+orig -epi_base 0 -anat2epi -anat_has_skull no -epi_strip 3dAutomask
|
(deprecated) anat+reg__ns_al_e2a_only_mat.aff12.1D |
The alignment/registration matrix of the anatomical data to the ASL Data. Produced from AFNI’s align_epi_anat.py. |
align_epi_anat.py -anat anat+reg__ns+orig -epi csfbriktcat1+reg+orig -epi_base 0 -anat2epi -anat_has_skull no -epi_strip 3dAutomask
|
anat+reg+orig |
The anatomical data aligned to the ASL data. |
3dAllineate -cubic -1Dmatrix_apply anat_ns_al_mat.aff12.1D -prefix anat+reg anat+orig
|
anat+reg+rs+orig |
Resampled version of anat+reg+orig to the ASL data voxel resolution. Produced from AFNI’s 3dResample. |
3dresample -rmode Cu -master aslbrik+dbltcat+reg+orig -prefix anat+reg+rs -inset anat+reg+orig
|
anat+reg+stripped+orig |
The anatomical data aligned to the ASL data and skull-stripped. Equivalent to anat_ns_al+orig |
3dAllineate -cubic -1Dmatrix_apply anat_ns_al_mat.aff12.1D -prefix anat+reg+stripped anat_ns+orig
|
anat+reg+stripped+rs+orig |
Resampled version of anat+reg+stripped+orig to the ASL data voxel resolution. Produced from AFNI’s 3dResample. |
3dresample -rmode Li -master aslbrik+dbltcat+reg+orig -prefix anat+reg+stripped+rs -inset anat+reg+stripped+orig
|
anat+reg+stripped+rs+mask+orig |
Brain mask derived from anat+reg+stripped+rs+orig |
Custom MATLAB code using thresholding of anat+reg+stripped+rs, smoothing, and imfill. |
anat+reg+betonreg+orig |
Skullstripped brain using FSL’s BET |
bet anat+reg.nii anat+reg+betonreg -f 0.3
3dcopy anat+reg+betonreg.nii anat+reg+betonreg
3drefit -orient RAI -duporigin anat+reg+orig anat+reg+betonreg+orig
|
anat+reg+betonreg+rs+orig |
Skullstripped brain using FSL’s BET, resampled to ASL grid. |
3dresample -rmode Li -master aslbrik+dbltcat+reg+orig -prefix anat+reg+betonreg+rs+orig -inset anat+reg+betonreg+orig
|
anat+reg+gray+pv+orig |
Gray matter partial volume data from using FSL’s FAST. |
fast -O 8 -g -o anat+reg+segonreg anat+reg+betonreg.nii
3dcopy anat+reg+segonreg_pve_1.nii anat+reg+gray+pv
3drefit -orient RAI -duporigin anat+reg+orig anat+reg+gray+pv+orig
|
anat+reg+gray+pv+rs+orig |
Gray matter partial volume data resampled to the ASL data voxel resolution, produced from AFNI’s 3dResample. |
3dresample -rmode Li -master aslbrik+dbltcat+reg+orig -prefix anat+reg+gray+pv+rs -inset anat+reg+gray+pv+orig
|
anat+reg+white+pv+orig |
White matter partial volume data from using FSL’s FAST. |
fast -O 8 -g -o anat+reg+segonreg anat+reg+betonreg.nii
3dcopy anat+reg+segonreg_pve_1.nii anat+reg+white+pv
3drefit -orient RAI -duporigin anat+reg+orig anat+reg+white+pv+orig
|
anat+reg+white+pv+rs+orig |
White matter partial volume data resampled to the ASL data voxel resolution, produced from AFNI’s 3dResample. |
3dresample -rmode Li -master aslbrik+dbltcat+reg+orig -prefix anat+reg+white+pv+rs -inset anat+reg+white+pv+orig
|
anat+reg+csf+pv+orig |
Cerebral spinal fluid partial volume data from using FSL’s FAST. |
fast -O 8 -g -o anat+reg+segonreg anat+reg+betonreg.nii
3dcopy anat+reg+segonreg_pve_1.nii anat+reg+csf+pv
3drefit -orient RAI -duporigin anat+reg+orig anat+reg+csf+pv+orig
|
anat+reg+csf+pv+rs+orig |
Cerebral spinal fluid partial volume data resampled to the ASL data voxel resolution, produced from AFNI’s 3dResample. |
3dresample -rmode Li -master aslbrik+dbltcat+reg+orig -prefix anat+reg+csf+pv+rs -inset anat+reg+csf+pv+orig
|
CBF+nomask+orig |
The cerebral blood flow (CBF) data calculated from the ASL and proton-density data. Units of mL/100g/min. |
Custom MATLAB code. Formula for CBF quantification depends on ASL pulse sequence and scan parameters used. |
CBF+orig |
The CBF data masked by anat+reg+stripped+rs+mask+orig. Units of mL/100g/min. |
Custom MATLAB code. Equivalent to CBF+nomask+orig masked by anat+reg+stripped+rs+mask+orig |
gm.txt |
Average gray matter CBF, units of mL/100g/min. Gray matter voxels picked out by thresholding the GM PV data. By default, the threshold is 0.6 to be considered a GM voxel. |
Custom MATLAB code. Equivalent to the following: 3dcalc -a anat+reg+gray+pv+rs+orig -b anat+reg+stripped+rs+mask+orig -expr 'ispositive(a-0.6)*b' -prefix temp
3dROIstats -mask temp+orig. CBF+nomask+orig.
|
white.txt |
Average white matter CBF, units of mL/100g/min. White matter voxels picked out by thresholding the WM PV data. By default, the threshold is 0.6 to be considered a WM voxel. |
Custom MATLAB code. Equivalent to the following: 3dcalc -a anat+reg+white+pv+rs+orig -b anat+reg+stripped+rs+mask+orig -expr 'ispositive(a-0.6)*b' -prefix temp
3dROIstats -mask temp+orig. CBF+nomask+orig.
|
CBF_gm+rs+orig |
Gray matter CBF image produced via partial volume correction (PVC). This only exists if the pvc option is used. |
Custom MATLAB code. |
CBF_wm+rs+orig |
White matter CBF image produced via partial volume correction (PVC). This only exists if the pvc option is used. |
Custom MATLAB code. |